I’ll create an environmental science example analyzing the relationship between CO2 emissions and global temperature changes, then visualize the data.
1 | import numpy as np |
Let me explain this environmental science example:
Problem Statement:
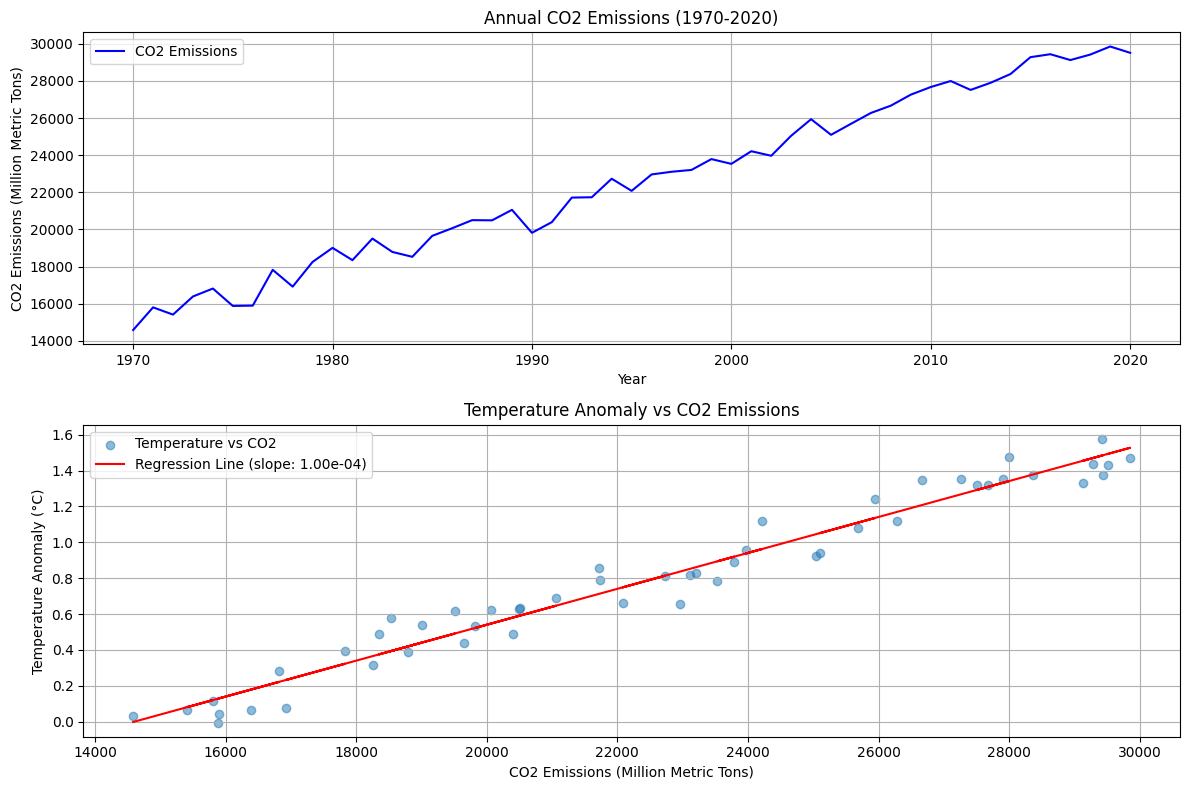
We’re analyzing the relationship between CO2 emissions and global temperature changes over a 50-year period (1970-2020).Data Simulation:
- We create synthetic data that mimics real-world patterns
- CO2 emissions show an increasing trend with random variations
- Temperature anomalies are modeled with a correlation to CO2 emissions
- Analysis Components:
- Correlation analysis between CO2 and temperature
- Linear regression to quantify the relationship
- Visualization of trends and relationships
- Visualization:
The code creates two plots:
- Top plot: Shows CO2 emissions trend over time
- Bottom plot: Displays the relationship between temperature anomalies and CO2 emissions, including a regression line
- Key Features:
- Error handling with random variations
- Statistical analysis (correlation coefficient and p-value)
- Clear visualization with proper labeling and grid lines
When you run this code, you’ll see:
- A clear upward trend in CO2 emissions over time
- A positive correlation between CO2 emissions and temperature anomalies
- Statistical measures of the relationship strength
Correlation coefficient: 0.981 P-value: 2.097e-36 Regression slope: 1.00e-04 °C/MtCO2