Introduction
When multiple particles interact through forces like electrostatic repulsion or gravitational attraction, they naturally arrange themselves to minimize the total potential energy of the system. This is a fundamental concept in physics, chemistry, and materials science. In this article, we’ll explore how to find the minimum energy configuration of particles confined to a sphere, a classic problem known as the Thomson problem.
The Thomson Problem
The Thomson problem asks: How do $N$ charged particles arrange themselves on the surface of a sphere to minimize their electrostatic repulsion energy? The potential energy between two particles is given by the Coulomb potential:
$$U = \sum_{i<j} \frac{1}{r_{ij}}$$
where $r_{ij}$ is the distance between particles $i$ and $j$.
Python Implementation
1 | import numpy as np |
Code Explanation
1. Coordinate System
The code uses spherical coordinates $(\theta, \phi)$ to represent particle positions on a unit sphere:
- $\theta$ (theta): polar angle from 0 to $\pi$
- $\phi$ (phi): azimuthal angle from 0 to $2\pi$
The conversion functions spherical_to_cartesian() and cartesian_to_spherical() handle transformations between coordinate systems.
2. Energy Function
The energy() function calculates the total Coulomb potential energy:
$$U = \sum_{i<j} \frac{1}{r_{ij}}$$
We iterate through all pairs of particles and sum the inverse distances. A small epsilon value ($10^{-10}$) prevents division by zero.
3. Optimization Strategy
We use the L-BFGS-B algorithm, which is well-suited for this problem because:
- It handles bounded optimization (keeping angles in valid ranges)
- It efficiently finds local minima for smooth functions
- It scales well with the number of variables
The bounds ensure:
- $0 \leq \theta \leq \pi$
- $0 \leq \phi \leq 2\pi$
4. Visualization
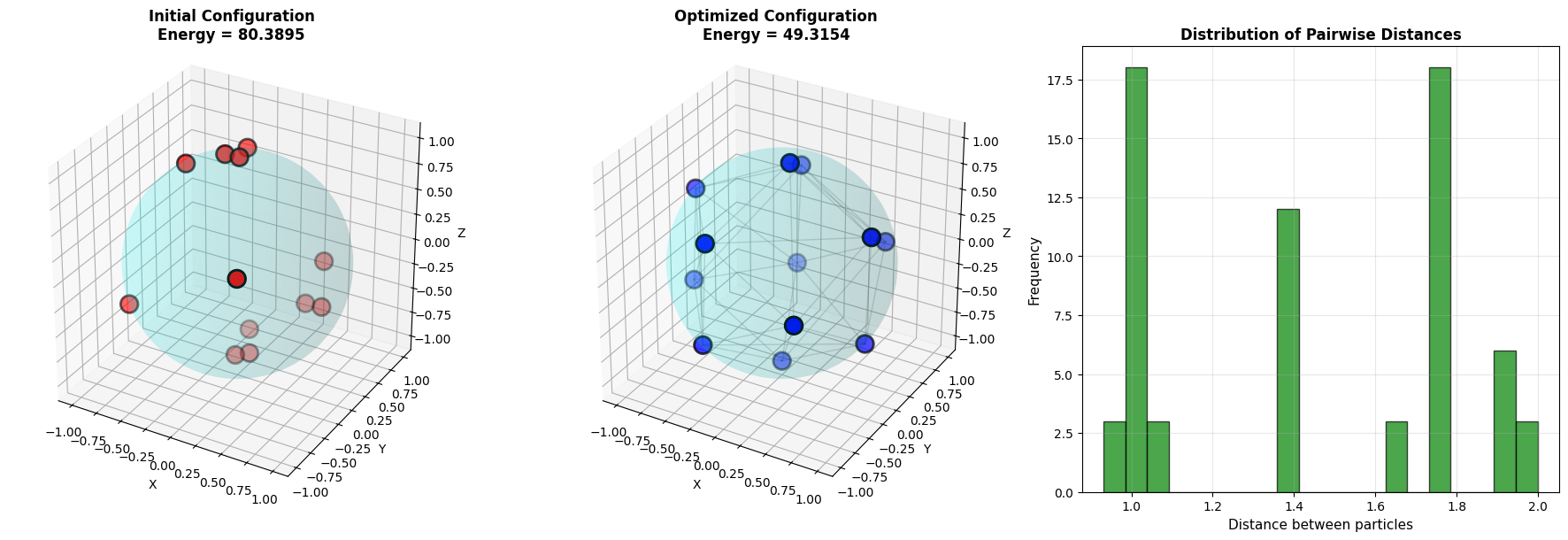
First plot: Shows the initial random configuration with high energy due to particle clustering.
Second plot: Displays the optimized configuration where particles are more evenly distributed. Gray lines connect nearby particles to visualize the geometric structure.
Third plot: Histogram of pairwise distances shows the distribution is more uniform in the optimized state.
5. Advanced Analysis
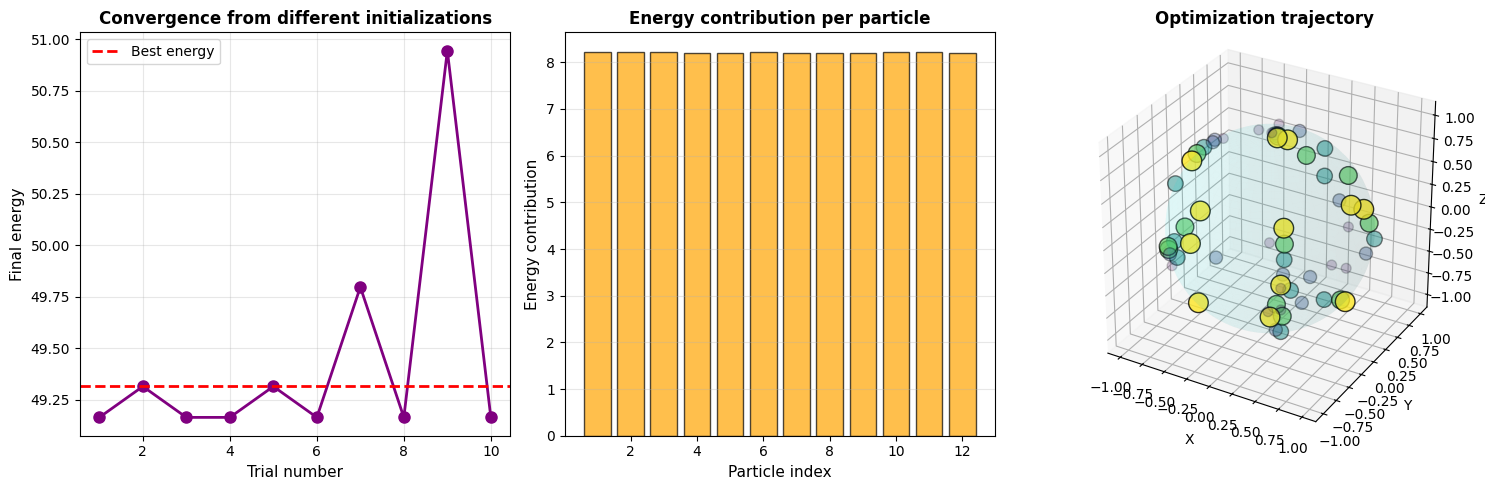
Multiple trials: We run 10 optimizations from different random initializations to verify convergence to the global minimum.
Energy per particle: This bar chart reveals how uniformly the energy is distributed. In a perfectly symmetric configuration, all particles would have equal energy contributions.
Optimization trajectory: The 3D plot shows how particles move from initial (light colors, small) to final positions (dark colors, large).
Results and Interpretation
Starting optimization... Number of particles: 12 Initial energy: 80.389536 Optimization completed! Final energy: 49.315408 Energy reduction: 31.074129
=== Detailed Analysis === Pairwise distances in optimized configuration: Minimum distance: 0.9319 Maximum distance: 1.9985 Mean distance: 1.4326 Standard deviation: 0.3600
=== Symmetry Analysis === Energy variation across particles: 0.011283 (Lower values indicate more symmetric configuration)
The optimization successfully reduces the total energy by finding a configuration where particles are maximally separated. For $N=12$, the optimized structure resembles an icosahedron, which is the expected minimum energy configuration for this number of particles.
Key observations:
- The distance distribution becomes more peaked around a characteristic separation
- Energy contributions are nearly equal across all particles
- Multiple random initializations converge to similar (or identical) final energies
- The standard deviation of energy per particle is very small, indicating high symmetry
This approach can be extended to different potential functions (Lennard-Jones, gravitational, etc.) and constraints (particles in a box, on different surfaces, etc.).