1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
31
32
33
34
35
36
37
38
39
40
41
42
43
44
45
46
47
48
49
50
51
52
53
54
55
56
57
58
59
60
61
62
63
64
65
66
67
68
69
70
71
72
73
74
75
76
77
78
79
80
81
82
83
84
85
86
87
88
89
90
91
92
93
94
95
96
97
98
99
100
101
102
103
104
105
106
107
108
109
110
111
112
113
114
115
116
117
118
119
120
121
122
123
124
125
126
127
128
129
130
131
132
133
134
135
136
137
138
139
140
141
142
143
144
145
146
147
148
149
150
151
152
153
154
155
156
157
158
159
160
161
162
163
164
165
166
167
168
169
170
171
172
173
174
175
176
177
178
179
180
181
182
183
184
185
186
187
188
189
190
191
192
193
194
195
196
197
198
199
200
201
202
203
204
205
206
207
208
209
210
211
212
213
214
215
216
217
218
219
220
221
222
223
224
225
226
227
228
229
230
231
232
233
234
235
236
237
238
239
240
241
242
243
244
245
246
247
248
249
250
251
252
253
254
255
256
257
258
259
260
261
262
263
264
265
266
267
268
269
270
271
272
273
274
275
276
277
278
279
280
281
282
283
284
285
286
287
288
289
290
291
292
293
294
295
296
297
298
299
300
301
302
303
304
305
306
307
308
309
310
311
312
313
314
315
316
317
318
319
320
321
322
323
324
325
326
327
328
329
330
331
332
333
334
335
336
337
338
339
340
341
342
343
344
345
346
347
348
349
350
351
352
353
354
355
356
357
358
359
360
361
362
363
364
365
366
367
368
369
370
371
372
373
374
375
376
377
378
379
380
381
382
383
384
385
386
387
388
389
390
391
392
393
394
395
396
397
398
399
400
401
402
403
404
405
406
407
408
409
410
411
412
413
414
| import numpy as np
import matplotlib.pyplot as plt
from mpl_toolkits.mplot3d import Axes3D
from scipy.stats import beta, binom
from scipy.special import betaln
import seaborn as sns
sns.set_style("whitegrid")
plt.rcParams['figure.figsize'] = (12, 8)
plt.rcParams['font.size'] = 11
class BayesianLifeProbability:
"""
Bayesian estimation of life existence probability on exoplanets
"""
def __init__(self, prior_alpha, prior_beta):
"""
Initialize with prior Beta distribution parameters
Parameters:
-----------
prior_alpha : float
Alpha parameter of prior Beta distribution (successes + 1)
prior_beta : float
Beta parameter of prior Beta distribution (failures + 1)
"""
self.prior_alpha = prior_alpha
self.prior_beta = prior_beta
def prior_distribution(self, theta):
"""Calculate prior probability density"""
return beta.pdf(theta, self.prior_alpha, self.prior_beta)
def likelihood(self, theta, n_obs, n_detections):
"""
Calculate likelihood of observing n_detections in n_obs observations
Parameters:
-----------
theta : array-like
Probability values
n_obs : int
Number of observations
n_detections : int
Number of positive biosignature detections
"""
return binom.pmf(n_detections, n_obs, theta)
def posterior_distribution(self, theta, n_obs, n_detections):
"""
Calculate posterior probability density after observing data
Using conjugate prior property: Beta + Binomial = Beta
"""
post_alpha = self.prior_alpha + n_detections
post_beta = self.prior_beta + (n_obs - n_detections)
return beta.pdf(theta, post_alpha, post_beta)
def update_posterior_params(self, n_obs, n_detections):
"""Get updated posterior parameters"""
post_alpha = self.prior_alpha + n_detections
post_beta = self.prior_beta + (n_obs - n_detections)
return post_alpha, post_beta
def posterior_statistics(self, n_obs, n_detections):
"""Calculate posterior mean, mode, variance, and credible interval"""
post_alpha, post_beta = self.update_posterior_params(n_obs, n_detections)
mean = post_alpha / (post_alpha + post_beta)
if post_alpha > 1 and post_beta > 1:
mode = (post_alpha - 1) / (post_alpha + post_beta - 2)
else:
mode = None
variance = (post_alpha * post_beta) / \
((post_alpha + post_beta)**2 * (post_alpha + post_beta + 1))
std = np.sqrt(variance)
ci_lower = beta.ppf(0.025, post_alpha, post_beta)
ci_upper = beta.ppf(0.975, post_alpha, post_beta)
return {
'mean': mean,
'mode': mode,
'std': std,
'variance': variance,
'ci_95': (ci_lower, ci_upper)
}
def information_gain(self, n_obs, n_detections):
"""
Calculate Kullback-Leibler divergence from prior to posterior
Measures information gain from observations
"""
post_alpha, post_beta = self.update_posterior_params(n_obs, n_detections)
kl = betaln(self.prior_alpha, self.prior_beta) - \
betaln(post_alpha, post_beta) + \
(post_alpha - self.prior_alpha) * \
(np.euler_gamma + np.log(post_alpha + post_beta - 2) -
np.log(post_alpha - 1) if post_alpha > 1 else 0) + \
(post_beta - self.prior_beta) * \
(np.euler_gamma + np.log(post_alpha + post_beta - 2) -
np.log(post_beta - 1) if post_beta > 1 else 0)
return abs(kl)
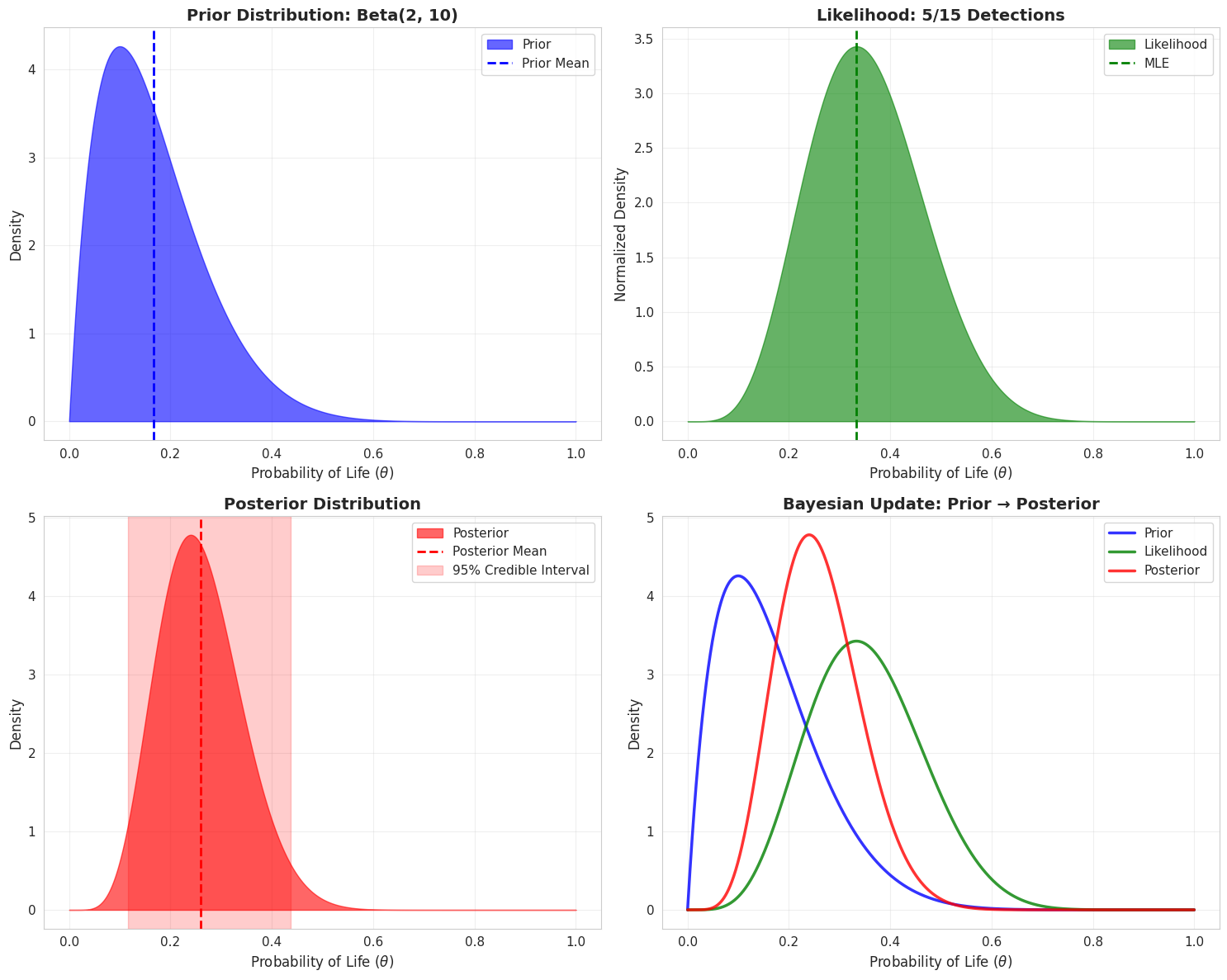
bayesian_model = BayesianLifeProbability(prior_alpha=2, prior_beta=10)
n_observations = 15
n_detections = 5
theta_values = np.linspace(0, 1, 1000)
prior = bayesian_model.prior_distribution(theta_values)
likelihood = bayesian_model.likelihood(theta_values, n_observations, n_detections)
posterior = bayesian_model.posterior_distribution(theta_values, n_observations, n_detections)
likelihood_normalized = likelihood / np.trapz(likelihood, theta_values)
stats = bayesian_model.posterior_statistics(n_observations, n_detections)
info_gain = bayesian_model.information_gain(n_observations, n_detections)
print("="*70)
print("BAYESIAN ESTIMATION OF LIFE EXISTENCE PROBABILITY")
print("="*70)
print(f"\nPrior Distribution: Beta(α={bayesian_model.prior_alpha}, β={bayesian_model.prior_beta})")
print(f"Prior Mean: {bayesian_model.prior_alpha/(bayesian_model.prior_alpha + bayesian_model.prior_beta):.4f}")
print(f"\nObservational Data:")
print(f" - Number of observations: {n_observations}")
print(f" - Positive detections: {n_detections}")
print(f" - Detection rate: {n_detections/n_observations:.2%}")
print(f"\nPosterior Statistics:")
print(f" - Mean: {stats['mean']:.4f}")
if stats['mode']:
print(f" - Mode: {stats['mode']:.4f}")
print(f" - Standard Deviation: {stats['std']:.4f}")
print(f" - 95% Credible Interval: [{stats['ci_95'][0]:.4f}, {stats['ci_95'][1]:.4f}]")
print(f"\nInformation Gain (KL Divergence): {info_gain:.4f}")
print(f"Uncertainty Reduction: {(1 - stats['std']/np.sqrt(bayesian_model.prior_alpha * bayesian_model.prior_beta /
((bayesian_model.prior_alpha + bayesian_model.prior_beta)**2 *
(bayesian_model.prior_alpha + bayesian_model.prior_beta + 1))))*100:.2f}%")
print("="*70)
fig, axes = plt.subplots(2, 2, figsize=(15, 12))
axes[0, 0].fill_between(theta_values, prior, alpha=0.6, color='blue', label='Prior')
axes[0, 0].axvline(bayesian_model.prior_alpha/(bayesian_model.prior_alpha + bayesian_model.prior_beta),
color='blue', linestyle='--', linewidth=2, label='Prior Mean')
axes[0, 0].set_xlabel(r'Probability of Life ($\theta$)', fontsize=12)
axes[0, 0].set_ylabel('Density', fontsize=12)
axes[0, 0].set_title('Prior Distribution: Beta(2, 10)', fontsize=14, fontweight='bold')
axes[0, 0].legend(fontsize=11)
axes[0, 0].grid(True, alpha=0.3)
axes[0, 1].fill_between(theta_values, likelihood_normalized, alpha=0.6, color='green', label='Likelihood')
axes[0, 1].axvline(n_detections/n_observations, color='green', linestyle='--',
linewidth=2, label='MLE')
axes[0, 1].set_xlabel(r'Probability of Life ($\theta$)', fontsize=12)
axes[0, 1].set_ylabel('Normalized Density', fontsize=12)
axes[0, 1].set_title(f'Likelihood: {n_detections}/{n_observations} Detections', fontsize=14, fontweight='bold')
axes[0, 1].legend(fontsize=11)
axes[0, 1].grid(True, alpha=0.3)
axes[1, 0].fill_between(theta_values, posterior, alpha=0.6, color='red', label='Posterior')
axes[1, 0].axvline(stats['mean'], color='red', linestyle='--', linewidth=2, label='Posterior Mean')
axes[1, 0].axvspan(stats['ci_95'][0], stats['ci_95'][1], alpha=0.2, color='red',
label='95% Credible Interval')
axes[1, 0].set_xlabel(r'Probability of Life ($\theta$)', fontsize=12)
axes[1, 0].set_ylabel('Density', fontsize=12)
axes[1, 0].set_title('Posterior Distribution', fontsize=14, fontweight='bold')
axes[1, 0].legend(fontsize=11)
axes[1, 0].grid(True, alpha=0.3)
axes[1, 1].plot(theta_values, prior, 'b-', linewidth=2.5, label='Prior', alpha=0.8)
axes[1, 1].plot(theta_values, likelihood_normalized, 'g-', linewidth=2.5, label='Likelihood', alpha=0.8)
axes[1, 1].plot(theta_values, posterior, 'r-', linewidth=2.5, label='Posterior', alpha=0.8)
axes[1, 1].set_xlabel(r'Probability of Life ($\theta$)', fontsize=12)
axes[1, 1].set_ylabel('Density', fontsize=12)
axes[1, 1].set_title('Bayesian Update: Prior → Posterior', fontsize=14, fontweight='bold')
axes[1, 1].legend(fontsize=11)
axes[1, 1].grid(True, alpha=0.3)
plt.tight_layout()
plt.savefig('bayesian_life_probability_2d.png', dpi=300, bbox_inches='tight')
plt.show()
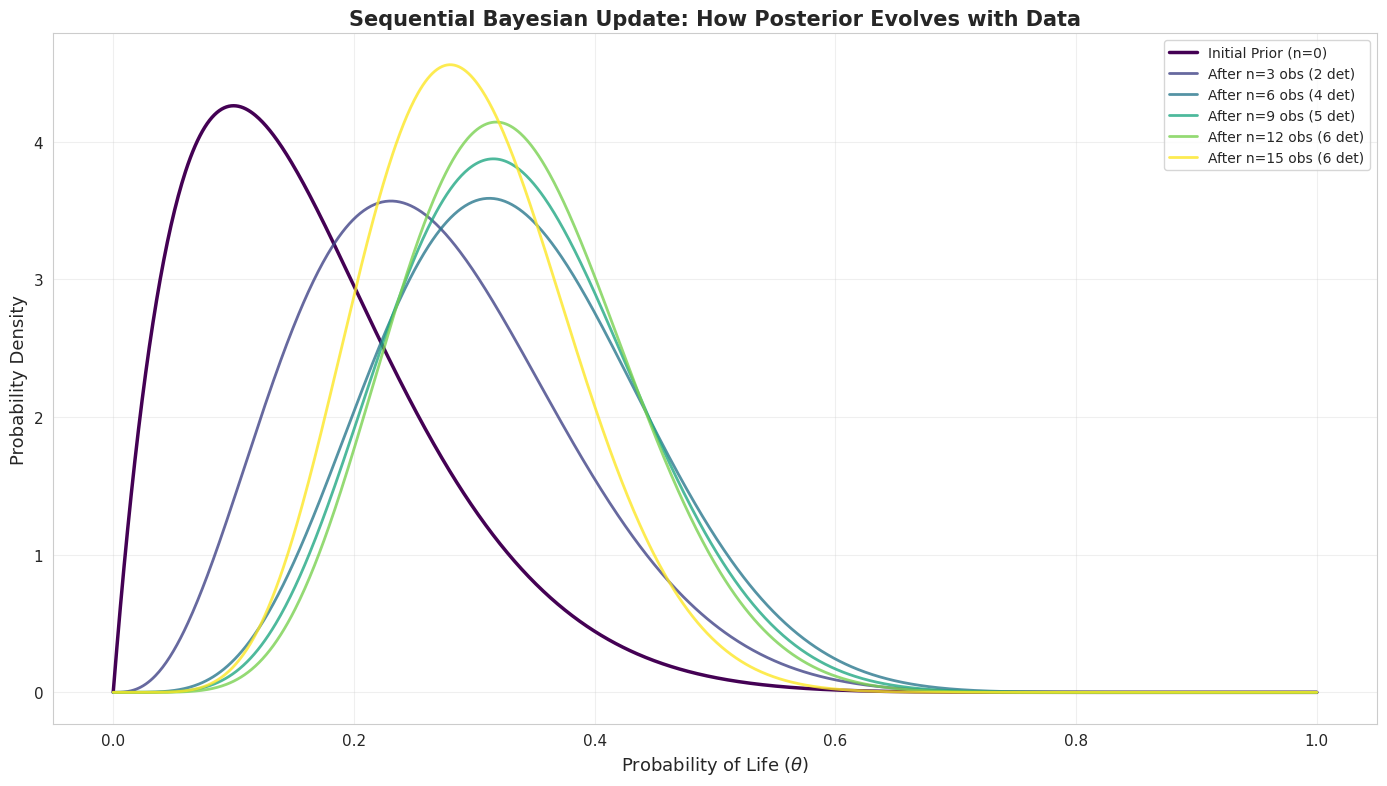
fig, ax = plt.subplots(figsize=(14, 8))
detections_sequence = [1, 0, 1, 0, 1, 1, 0, 0, 1, 0, 0, 1, 0, 0, 0]
colors = plt.cm.viridis(np.linspace(0, 1, len(detections_sequence) + 1))
current_alpha = bayesian_model.prior_alpha
current_beta = bayesian_model.prior_beta
ax.plot(theta_values, beta.pdf(theta_values, current_alpha, current_beta),
linewidth=2.5, label=f'Initial Prior (n=0)', color=colors[0])
cumulative_detections = 0
for i, detection in enumerate(detections_sequence):
cumulative_detections += detection
current_alpha += detection
current_beta += (1 - detection)
if (i + 1) % 3 == 0 or i == len(detections_sequence) - 1:
ax.plot(theta_values, beta.pdf(theta_values, current_alpha, current_beta),
linewidth=2, label=f'After n={i+1} obs ({cumulative_detections} det)',
color=colors[i+1], alpha=0.8)
ax.set_xlabel(r'Probability of Life ($\theta$)', fontsize=13)
ax.set_ylabel('Probability Density', fontsize=13)
ax.set_title('Sequential Bayesian Update: How Posterior Evolves with Data',
fontsize=15, fontweight='bold')
ax.legend(fontsize=10, loc='upper right')
ax.grid(True, alpha=0.3)
plt.tight_layout()
plt.savefig('sequential_bayesian_update.png', dpi=300, bbox_inches='tight')
plt.show()
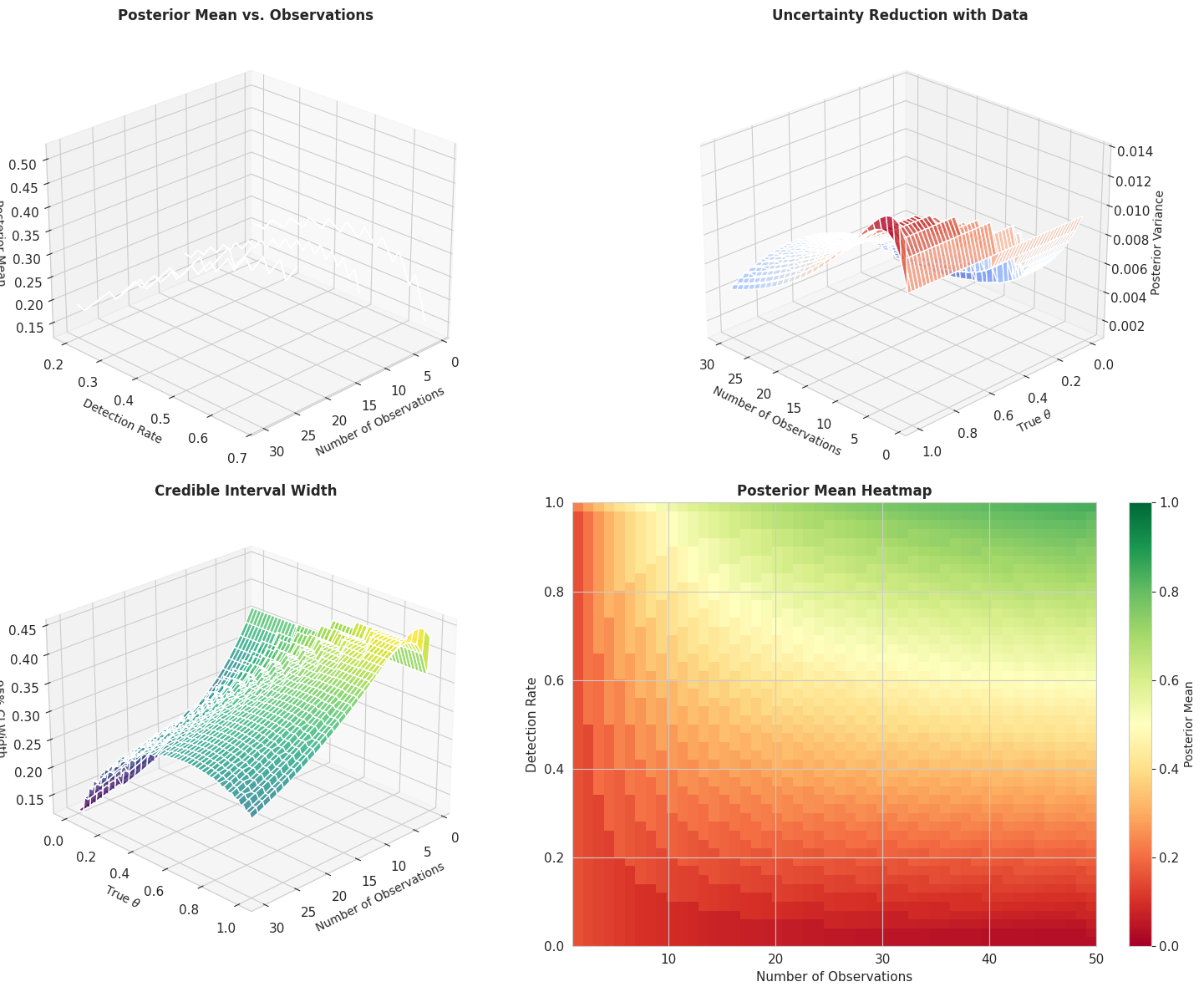
fig = plt.figure(figsize=(16, 12))
ax1 = fig.add_subplot(221, projection='3d')
ax2 = fig.add_subplot(222, projection='3d')
ax3 = fig.add_subplot(223, projection='3d')
ax4 = fig.add_subplot(224)
n_obs_range = np.arange(1, 31)
theta_range = np.linspace(0, 1, 100)
N_OBS, THETA = np.meshgrid(n_obs_range, theta_range)
detection_ratios = [0.2, 0.33, 0.5, 0.67]
for ratio in detection_ratios:
posterior_means = np.zeros_like(N_OBS, dtype=float)
for i, n in enumerate(n_obs_range):
n_det = int(n * ratio)
post_alpha = bayesian_model.prior_alpha + n_det
post_beta = bayesian_model.prior_beta + (n - n_det)
mean = post_alpha / (post_alpha + post_beta)
posterior_means[:, i] = mean
ax1.plot_surface(N_OBS, THETA * 0 + ratio, posterior_means, alpha=0.4,
label=f'Detection rate={ratio:.0%}')
ax1.set_xlabel('Number of Observations', fontsize=10)
ax1.set_ylabel('Detection Rate', fontsize=10)
ax1.set_zlabel('Posterior Mean', fontsize=10)
ax1.set_title('Posterior Mean vs. Observations', fontsize=12, fontweight='bold')
ax1.view_init(elev=25, azim=45)
Z_variance = np.zeros_like(N_OBS, dtype=float)
for i, n in enumerate(n_obs_range):
for j, theta in enumerate(theta_range):
n_det = int(n * theta)
post_alpha = bayesian_model.prior_alpha + n_det
post_beta = bayesian_model.prior_beta + (n - n_det)
variance = (post_alpha * post_beta) / \
((post_alpha + post_beta)**2 * (post_alpha + post_beta + 1))
Z_variance[j, i] = variance
surf2 = ax2.plot_surface(N_OBS, THETA, Z_variance, cmap='coolwarm', alpha=0.8)
ax2.set_xlabel('Number of Observations', fontsize=10)
ax2.set_ylabel(r'True $\theta$', fontsize=10)
ax2.set_zlabel('Posterior Variance', fontsize=10)
ax2.set_title('Uncertainty Reduction with Data', fontsize=12, fontweight='bold')
ax2.view_init(elev=25, azim=135)
Z_ci_width = np.zeros_like(N_OBS, dtype=float)
for i, n in enumerate(n_obs_range):
for j, theta in enumerate(theta_range):
n_det = int(n * theta)
post_alpha = bayesian_model.prior_alpha + n_det
post_beta = bayesian_model.prior_beta + (n - n_det)
ci_lower = beta.ppf(0.025, post_alpha, post_beta)
ci_upper = beta.ppf(0.975, post_alpha, post_beta)
Z_ci_width[j, i] = ci_upper - ci_lower
surf3 = ax3.plot_surface(N_OBS, THETA, Z_ci_width, cmap='viridis', alpha=0.8)
ax3.set_xlabel('Number of Observations', fontsize=10)
ax3.set_ylabel(r'True $\theta$', fontsize=10)
ax3.set_zlabel('95% CI Width', fontsize=10)
ax3.set_title('Credible Interval Width', fontsize=12, fontweight='bold')
ax3.view_init(elev=25, azim=45)
n_obs_heatmap = np.arange(1, 51)
theta_heatmap = np.linspace(0, 1, 50)
posterior_mean_grid = np.zeros((len(theta_heatmap), len(n_obs_heatmap)))
for i, n in enumerate(n_obs_heatmap):
for j, theta in enumerate(theta_heatmap):
n_det = int(n * theta)
post_alpha = bayesian_model.prior_alpha + n_det
post_beta = bayesian_model.prior_beta + (n - n_det)
posterior_mean_grid[j, i] = post_alpha / (post_alpha + post_beta)
im = ax4.imshow(posterior_mean_grid, aspect='auto', origin='lower',
extent=[1, 50, 0, 1], cmap='RdYlGn', vmin=0, vmax=1)
ax4.set_xlabel('Number of Observations', fontsize=11)
ax4.set_ylabel('Detection Rate', fontsize=11)
ax4.set_title('Posterior Mean Heatmap', fontsize=12, fontweight='bold')
cbar = plt.colorbar(im, ax=ax4)
cbar.set_label('Posterior Mean', fontsize=10)
plt.tight_layout()
plt.savefig('bayesian_life_probability_3d.png', dpi=300, bbox_inches='tight')
plt.show()
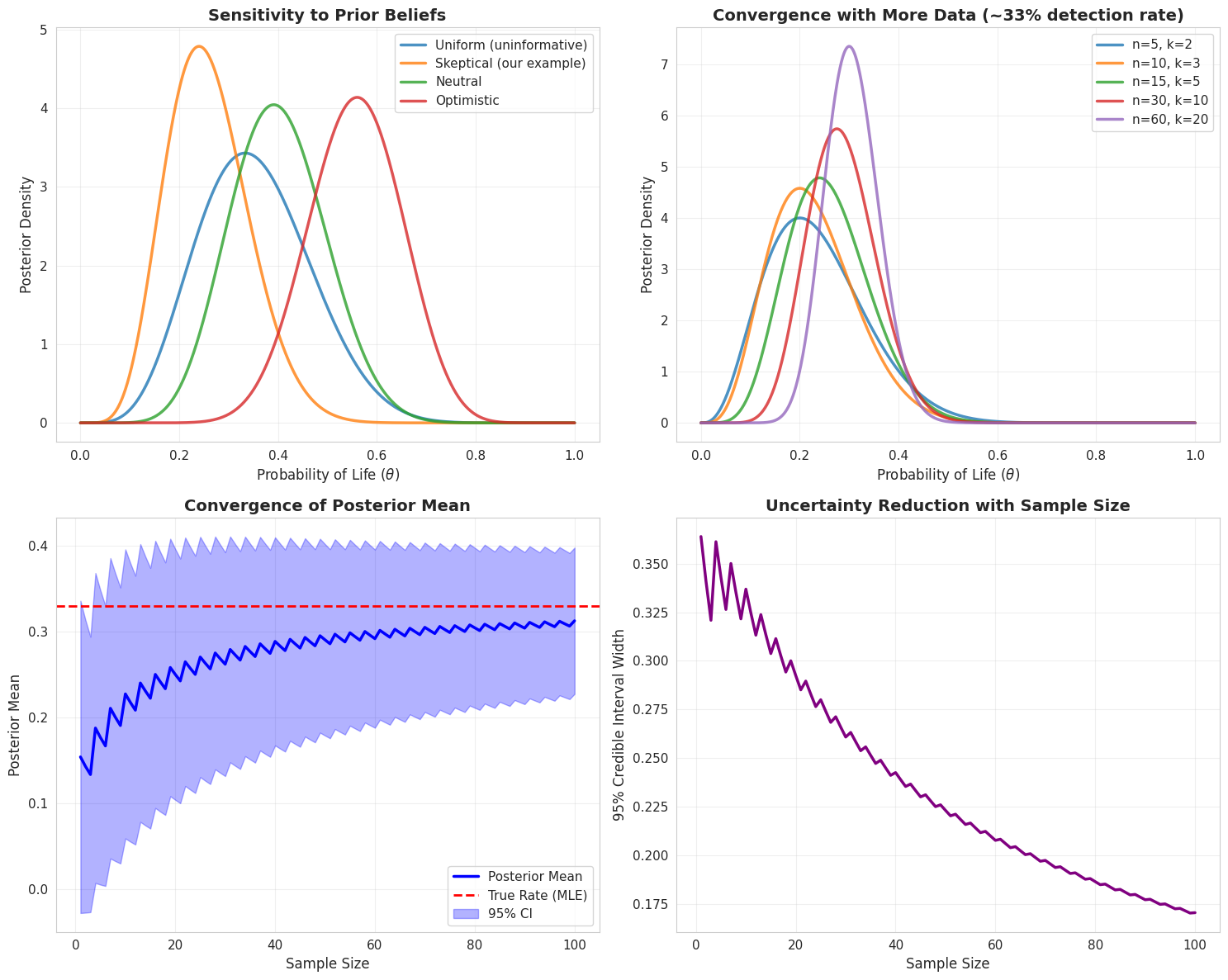
fig, axes = plt.subplots(2, 2, figsize=(15, 12))
priors_to_test = [
(1, 1, "Uniform (uninformative)"),
(2, 10, "Skeptical (our example)"),
(5, 5, "Neutral"),
(10, 2, "Optimistic")
]
for prior_params in priors_to_test:
alpha, beta_param, label = prior_params
model = BayesianLifeProbability(alpha, beta_param)
posterior = model.posterior_distribution(theta_values, n_observations, n_detections)
axes[0, 0].plot(theta_values, posterior, linewidth=2.5, label=label, alpha=0.8)
axes[0, 0].set_xlabel(r'Probability of Life ($\theta$)', fontsize=12)
axes[0, 0].set_ylabel('Posterior Density', fontsize=12)
axes[0, 0].set_title('Sensitivity to Prior Beliefs', fontsize=14, fontweight='bold')
axes[0, 0].legend(fontsize=11)
axes[0, 0].grid(True, alpha=0.3)
data_amounts = [(5, 2), (10, 3), (15, 5), (30, 10), (60, 20)]
for n, k in data_amounts:
posterior = bayesian_model.posterior_distribution(theta_values, n, k)
axes[0, 1].plot(theta_values, posterior, linewidth=2.5,
label=f'n={n}, k={k}', alpha=0.8)
axes[0, 1].set_xlabel(r'Probability of Life ($\theta$)', fontsize=12)
axes[0, 1].set_ylabel('Posterior Density', fontsize=12)
axes[0, 1].set_title('Convergence with More Data (~33% detection rate)',
fontsize=14, fontweight='bold')
axes[0, 1].legend(fontsize=11)
axes[0, 1].grid(True, alpha=0.3)
sample_sizes = np.arange(1, 101)
posterior_means = []
ci_widths = []
for n in sample_sizes:
k = int(n * 0.33)
stats = bayesian_model.posterior_statistics(n, k)
posterior_means.append(stats['mean'])
ci_widths.append(stats['ci_95'][1] - stats['ci_95'][0])
axes[1, 0].plot(sample_sizes, posterior_means, 'b-', linewidth=2.5, label='Posterior Mean')
axes[1, 0].axhline(0.33, color='red', linestyle='--', linewidth=2, label='True Rate (MLE)')
axes[1, 0].fill_between(sample_sizes,
np.array(posterior_means) - np.array(ci_widths)/2,
np.array(posterior_means) + np.array(ci_widths)/2,
alpha=0.3, color='blue', label='95% CI')
axes[1, 0].set_xlabel('Sample Size', fontsize=12)
axes[1, 0].set_ylabel('Posterior Mean', fontsize=12)
axes[1, 0].set_title('Convergence of Posterior Mean', fontsize=14, fontweight='bold')
axes[1, 0].legend(fontsize=11)
axes[1, 0].grid(True, alpha=0.3)
axes[1, 1].plot(sample_sizes, ci_widths, 'purple', linewidth=2.5)
axes[1, 1].set_xlabel('Sample Size', fontsize=12)
axes[1, 1].set_ylabel('95% Credible Interval Width', fontsize=12)
axes[1, 1].set_title('Uncertainty Reduction with Sample Size', fontsize=14, fontweight='bold')
axes[1, 1].grid(True, alpha=0.3)
plt.tight_layout()
plt.savefig('sensitivity_analysis.png', dpi=300, bbox_inches='tight')
plt.show()
print("\nVisualization complete! All graphs have been generated.")
|